|
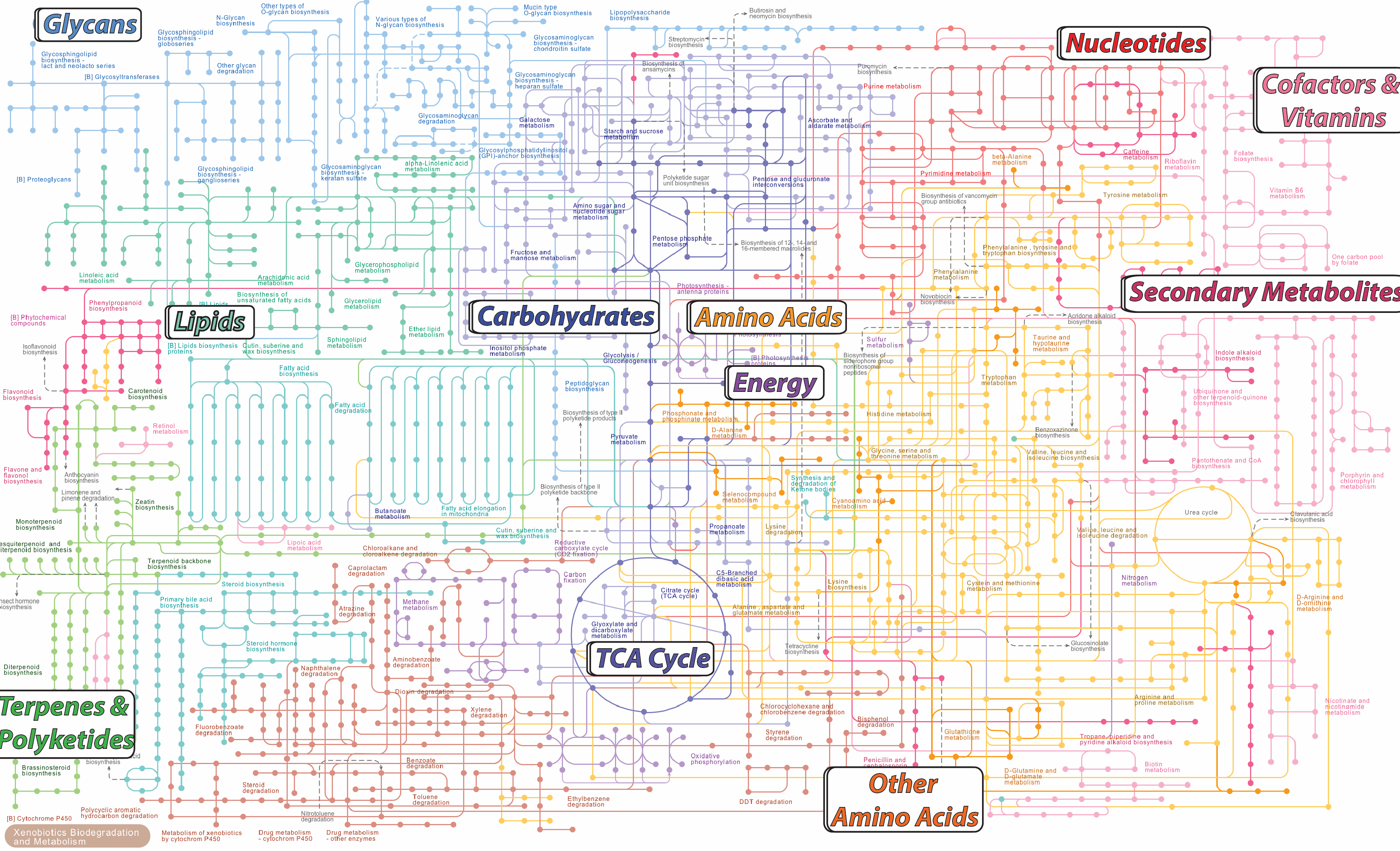
MetabolomicsMetabolomics is the science and technology of the measurement, identification, and quantification of metabolites in living systems. Metabolites are sensitive to genetics and the environment, and they are closely related to behavioral phenotypes. In a systems biology analysis, metabolomics can provide important data that can improve the understanding and interpretation of other omics such as genomics, transcriptomics, and proteomics. |
|
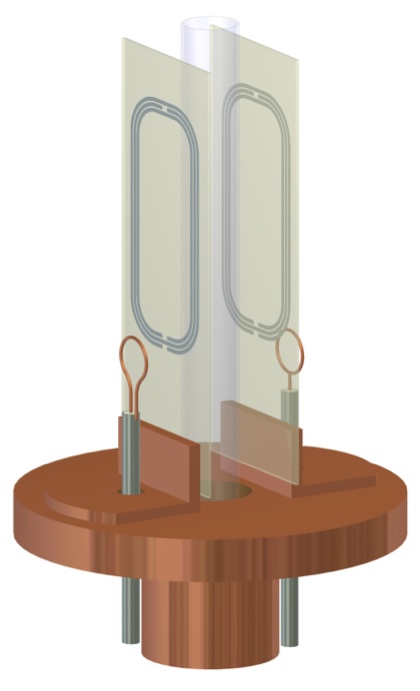
High Sensitivity NMR Probe DevelopmentNMR is inherently very insensitive but it is one of the most important analytical techniques for metabolomics, natural product discovery and structural biology. To improve the situation, we have worked with engineering groups at the National High Magnetic Field Laboratory and elsewhere in the world to develop new NMR probes that will enhance applications in protein and natural product studies. We previously developed a unique 1-mm high temperature superconducting (HTS) probe with Bruker and the NHMFL (Bill Brey, Rich Withers, and Rob Nast). Although no longer operational, the 1-mm HTS probe is one of the most mass sensitive probes in the world. These probes have coils that are made from thin films of YBCO deposited onto sapphire substrates to achieve very high Q. |
|
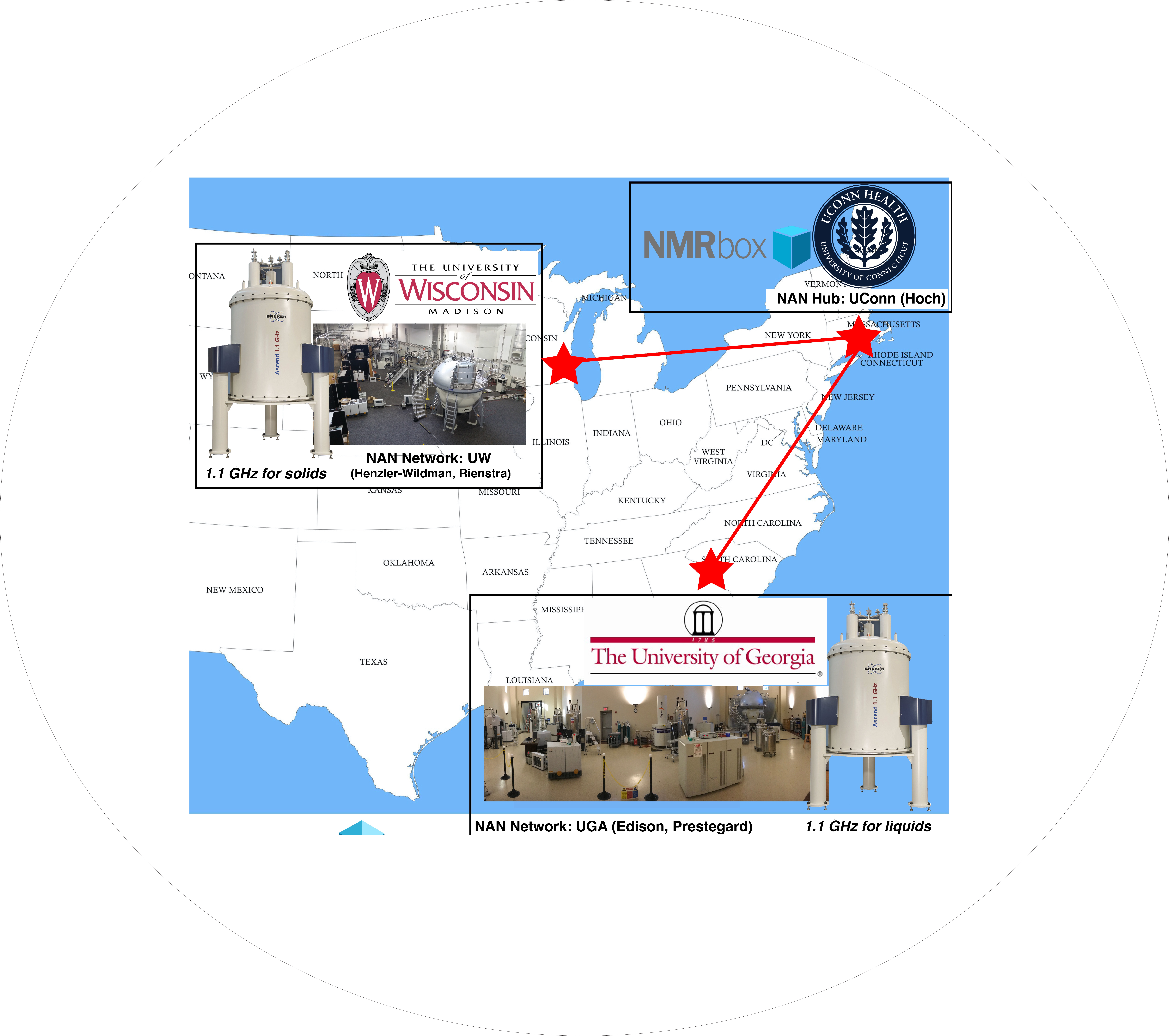
Network for Advanced NMRThe Network for Advanced NMR (NAN) is supported by the NSF Mid-scale Research Infrastructure-2 program. UGA partners with the University of Connecticut Health Center and NMRFAM at the University of Wisconsin-Madison. UGA will get a 1.1 GHz NMR that will be dedicated to biological solution NMR. UW-Madison will get a 1.1 GHz NMR dedicated to biosolids and materials. UConn is the central hub, where data are automatically and securely stored. Users will be able to use “knowledgebases” in 4 areas: 1) Metabolomics, 2) Solution structural biology, 3) Solid-state structural biology, and 4) Materials. UGA is developing the first two knowledgebases. The knowledgebases include NMR protocols and pulse sequences with example datasets, training videos and other training material, and data processing and analysis protocols. Much of the protocol information is being shared through protocols.io. Users will also be able to discover instruments that would be best for their study through a discovery portal that is modeled loosly like airBnB (e.g, airB0). Experienced users will be able to send samples to NAN facilities and operate the instruments remotely. Data are transfered to NMRbox, which is operated by the NAN collaborators at UConn. One of our primary goals is to make NMR more accessible to non-experts: The Democratization of NMR. I am trying to make some videos that explain NMR to non-experts. This project is going slowly, but the first in the series is available on YouTube. Feedback is welcome! |
|
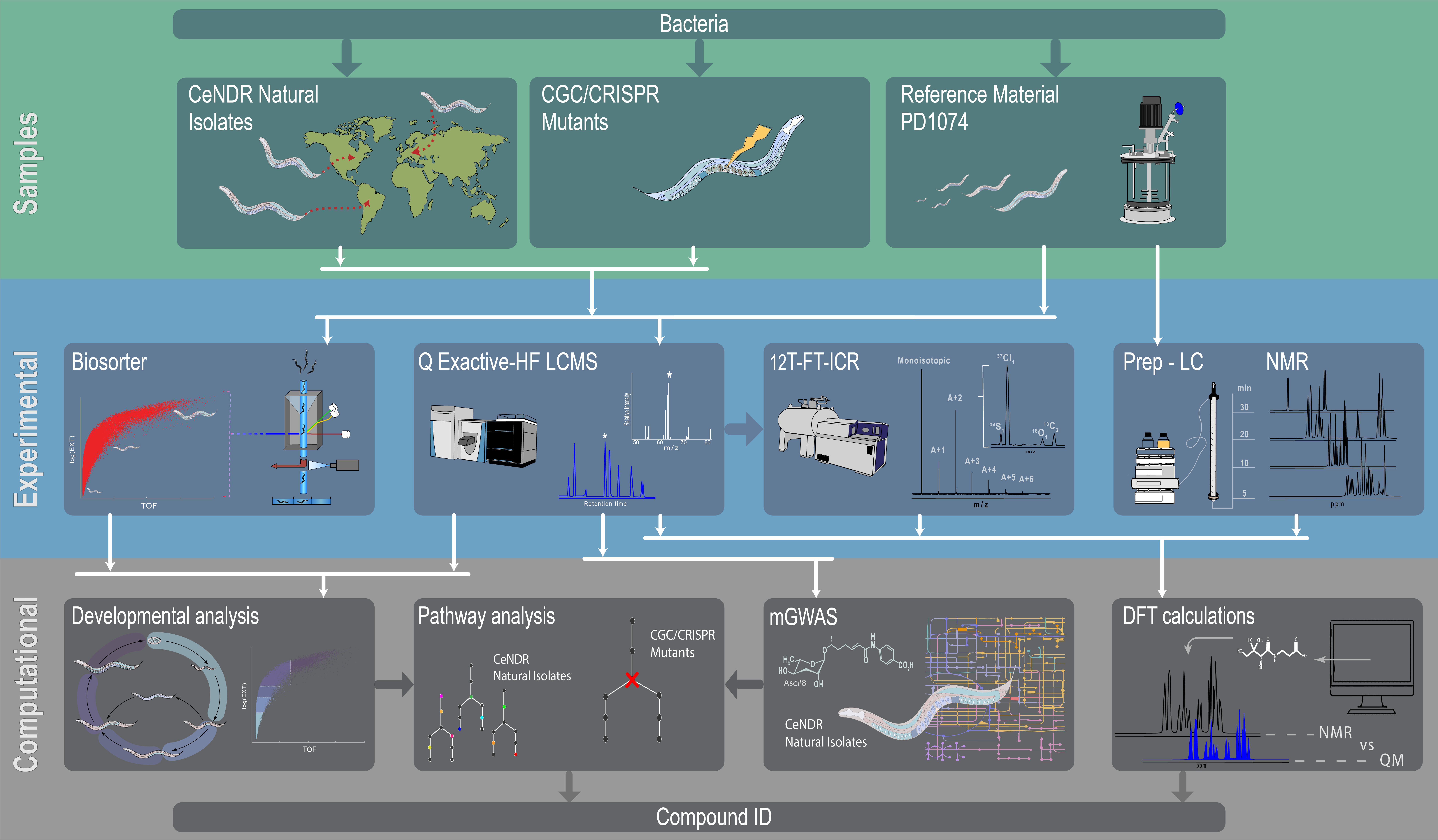
Unknown Compound IdentificationUnknown compound identification is a major challenge in metabolomics. Even establishing broad definitions of an “unknown compound” is not straightforward. An unlikely source of clarification (some might say confusion) to our field came from former US Defense Sectretary, Donald Rumsfeld, who once said
Following Rumsfeld, a “known known” is a metabolite in a reference database that can be matched with an experimental dataset. A “known unknown” is a metabolite that has been found before but is not in an accessible database. An “unknown unknown” is a metabolite that has not been discovered. The NIH Metabolomics Common Fund has funded 5 centers in the US to improve unknown metabolite identification. Our lab leads one of these centers, and our project is titled Genetics and Quantum Chemistry as Tools for Unknown Metabolite Identification. We are using the model organism Caenorhabditis elegans and comparing both known mutants and natural isolates with the reference strain PD1074. We collaborate with Erik Anderson on C. elegans and Lauren McIntyre on study design and biostatistics. The conceptual steps that we use in this project are:
|
|
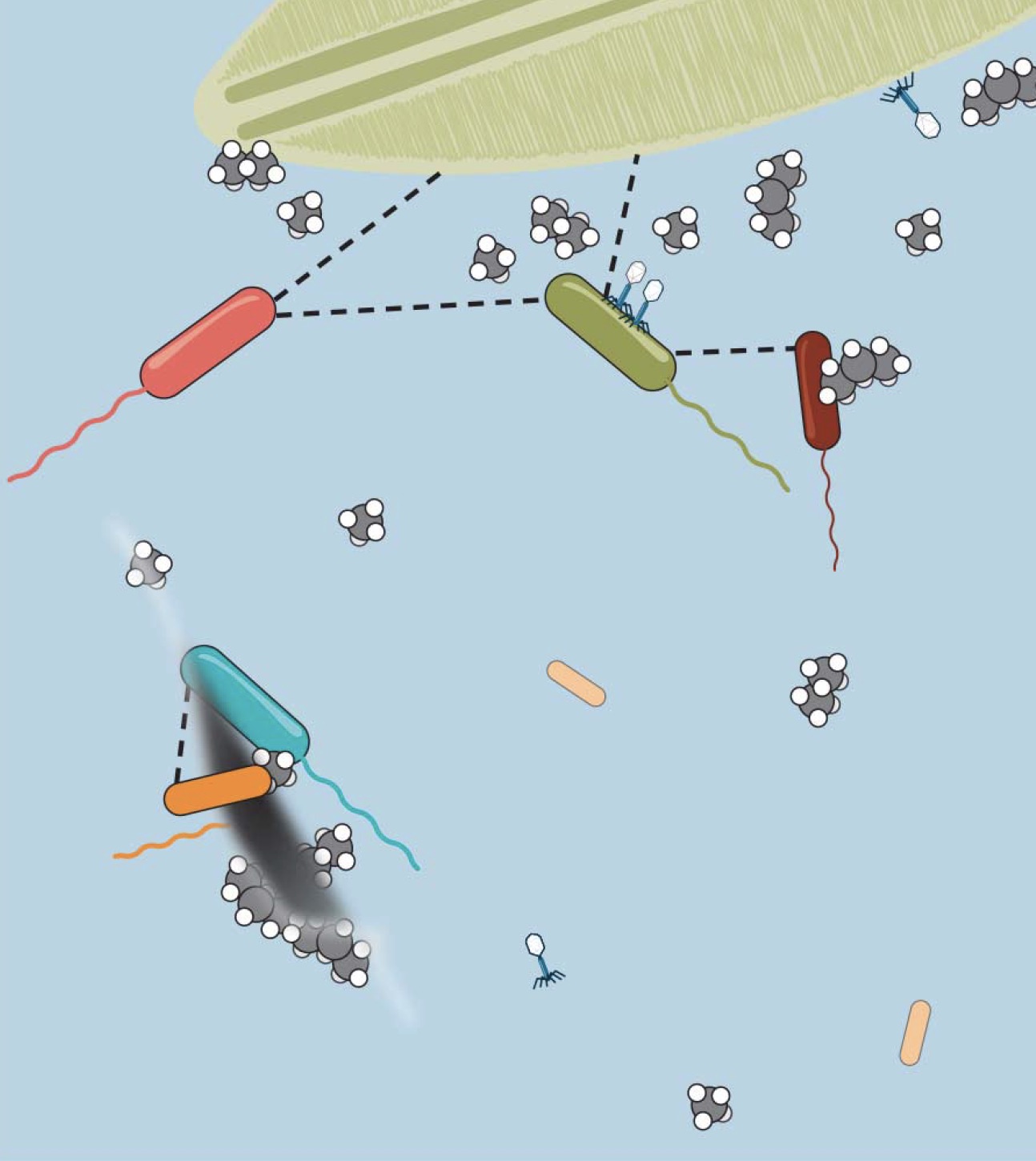
Carbon Cycling in the OceanDid you know that half of the oxygen that you are breathing is produced by phytoplankton in the ocean? We are studying fast carbon cycling in the shallow ocean with an amazing team of scientists who are marine scientists, analytical chemists, microbiologists, virologists, data scientists, and modelers. It is a wonderful interdisciplinary group! We are funded by an NSF Science and Technology Center, and we are called the “Center for Chemical Currencies of a Microbial Planet (C-CoMP)”. Our role in C-CoMP is to help measure and quantify small metabolites that are produced by phytoplankton and released to marine heterotrophic bacteria. About 50% of the CO2 in the earth’s atmosphere cycles through the shallow ocean in about 1 week! The phytoplankton photosynthesize the CO2 and release about half of that carbon to bacteria, which are part of a huge ocean microbiome. These bacteria release CO2 back into the atmosphere through respiration, and they form the basis of the marine food chain. This carbon cycle is clearly very important to earth, but very little is known about its dependence on climate change, especially warmer and more acidic oceans. There are some challenges in measuring these small metabolites in the carbon cycle. First, they are very dilute and need concentration for NMR or mass spectrometry. Second, they are very hard to separate from salt, which is at high concentrations in the ocean. But the salt makes the NMR and MS measurements challenging. We are working on a number of different approaches to improve this situation. Please see the publication list for some articles related to this topic. C-CoMP team members wrote a review article that is a nice place to learn more. |
|
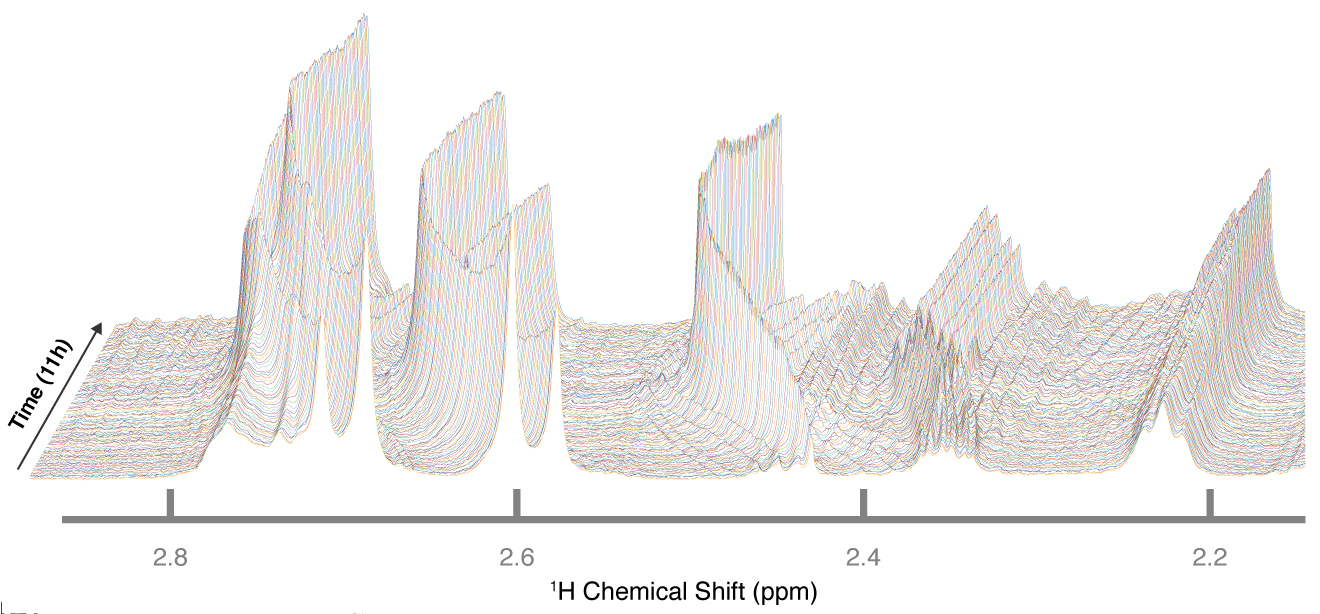
ClockWe collaborate with Prof. Jonathan Arnold and others to try to understand the circadian oscillation of the filamentous fungus, Neurospora crassa. This model organism has a clock that cycles about every 24 hours, and our lab is trying to understand the chemical control of that behavior. N. crassa is also a famous model organism because of the Nobel Prize research by Beadle and Tatum. In that work, Beadle and Tatum discovered a way to link metabolism with genetics, which has been credited as the start of We collaborate with Prof. Jonathan Arnold and others to try to understand the circadian oscillation of the filamentous fungus, Neurospora crassa. This model organism has a clock that cycles about every 24 hours, and our lab is trying to understand the chemical control of that behavior. N. crassa is also a famous model organism because of the Nobel Prize research by Beadle and Tatum. In that work, Beadle and Tatum discovered a way to link metabolism with genetics, which has been credited as the start of molecular biology. Our research into the clock of N. crassa brought us to a new method that we have develped called Continous in vivo Metabolism by NMR (CIVM-NMR). Using CIVM-NMR, we can extract detailed information on the changes in metabolites as the orgamism grows. We can study different genetic strains of N. crassa and different sources of nutrients or oxygenation levels. |

|
Dog NutritionWe collaborate with Prof. Joseph Bartges in the UGA vet school. Joe is an expert in clinical nutrition and is especially interested in advanced glycation end products (AGEs), which are present at high abundance in processed food, such as canned dog food or kibble. AGEs cause a number of health problems in people and pets, and we are trying to help Joe and his team understand the relationship between AGEs and metabolites. |

|
Cell based therapeuticsUnder construction |

|
Stress in PregnacyWe are no longer working on this project. |